| USPEX 10.5 manual |
These are stored in the folder  /StructurePrediction/results1. If this is a new calculation, results2, results3, …(if the calculation has been restarted or run a few times), there will be a separate results* folder for each calculation. Caution: When looking at space groups in the file Individuals, keep in mind that USPEX often underdetermines space group symmetries, because of finite precision of structure relaxation and relatively tight space group determination tolerances. You should visualize the predicted structures. To get full space group, either increase symmetry tolerances (but this can be dangerous), or re-relax your structure with increased precision.
/StructurePrediction/results1. If this is a new calculation, results2, results3, …(if the calculation has been restarted or run a few times), there will be a separate results* folder for each calculation. Caution: When looking at space groups in the file Individuals, keep in mind that USPEX often underdetermines space group symmetries, because of finite precision of structure relaxation and relatively tight space group determination tolerances. You should visualize the predicted structures. To get full space group, either increase symmetry tolerances (but this can be dangerous), or re-relax your structure with increased precision.
The subdirectory  /StructurePrediction/results1 contains the following files:
/StructurePrediction/results1 contains the following files:
OUTPUT.txt — summarizes input variables, structures produced by USPEX, and their characteristics.
Parameters.txt — this is a copy of the INPUT.txt file used in this calculation, for your reference.
Individuals — gives details of all produced structures (energies, unit cell volumes, space groups, variation operators that were used to produce the structures, 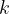 -points mesh used to compute the structures’ final energy, degrees of order, etc.). File BESTIndividuals gives this information for the best structures from each generation. Example of Individuals file:
-points mesh used to compute the structures’ final energy, degrees of order, etc.). File BESTIndividuals gives this information for the best structures from each generation. Example of Individuals file:
Gen ID Origin Composition Enthalpy Volume Density Fitness KPOINTS SYMM Q_entr A_order S_order
(eV) (A^3) (g/cm^3)
1 1 Random [ 4 8 16 ] -655.062 201.062 4.700 -655.062 [ 1 1 1] 1 0.140 1.209 2.632
1 2 Random [ 4 8 16 ] -650.378 206.675 4.572 -650.378 [ 1 1 1] 1 0.195 1.050 2.142
1 3 Random [ 4 8 16 ] -646.184 203.354 4.647 -646.184 [ 1 1 1] 1 0.229 0.922 1.746
1 4 Random [ 4 8 16 ] -649.459 198.097 4.770 -649.459 [ 1 1 1] 9 0.128 0.958 2.171
1 5 Random [ 4 8 16 ] -648.352 202.711 4.662 -648.352 [ 1 1 1] 2 0.154 1.014 2.148
1 6 Random [ 4 8 16 ] -643.161 206.442 4.577 -643.161 [ 1 1 1] 1 0.234 0.946 1.766
1 7 Random [ 4 8 16 ] -647.678 207.119 4.562 -647.678 [ 1 1 1] 1 0.224 1.108 2.106
1 8 Random [ 4 8 16 ] -644.482 203.844 4.636 -644.482 [ 1 1 1] 1 0.215 0.952 1.857
1 9 Random [ 4 8 16 ] -647.287 204.762 4.615 -647.287 [ 1 1 1] 40 0.136 1.142 2.563
1 10 Random [ 4 8 16 ] -649.459 198.097 4.770 -649.459 [ 1 1 1] 9 0.128 0.958 2.171
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
convex_hull — only for variable-composition calculations, this file gives all thermodynamically stable compositions, and their enthalpies (per atom). Example:
---- generation 1 -------
10 0 -8.5889
0 14 -8.5893
11 3 -8.7679
---- generation 2 -------
10 0 -8.5889
0 14 -8.5893
11 3 -8.8204
---- generation 3 -------
10 0 -8.5889
0 14 -8.5893
12 4 -8.9945
. . . . . . . . . . . . .
gatheredPOSCARS — relaxed structures (in the VASP5 POSCAR format). Example:
EA1 9.346 8.002 2.688 90.000 90.000 90.000 Sym.group: 1
1.0000
9.346156 0.000000 0.000000
0.000000 8.002181 0.000000
0.000000 0.000000 2.688367
Mg Al O
4 8 16
Direct
0.487956 0.503856 0.516443
0.777565 0.007329 0.016443
0.987956 0.507329 0.016443
0.277565 0.003856 0.516443
0.016944 0.178753 0.016443
0.019294 0.833730 0.516443
0.746227 0.333730 0.516443
0.748577 0.678753 0.016443
0.516944 0.832431 0.516443
0.519294 0.177455 0.016443
0.246227 0.677455 0.016443
0.248577 0.332431 0.516443
0.416676 0.241774 0.516443
0.559871 0.674713 0.016443
0.205650 0.174713 0.016443
0.348845 0.741774 0.516443
0.613957 0.380343 0.016443
0.804054 0.542164 0.516443
0.113957 0.630842 0.516443
0.304054 0.469021 0.016443
0.848845 0.269411 0.016443
0.705650 0.836472 0.516443
0.059871 0.336472 0.516443
0.916676 0.769411 0.016443
0.651564 0.130842 0.516443
0.461467 0.969021 0.016443
0.151564 0.880343 0.016443
0.961467 0.042164 0.516443
EA2 9.487 4.757 4.580 90.243 90.188 89.349 Sym.group: 1
1.0000
9.486893 0.000000 0.000000
0.054041 4.756769 0.000000
-0.014991 -0.019246 4.579857
Mg Al O
4 8 16
Direct
0.499837 0.633752 0.011361
0.500082 0.131390 0.482012
0.813573 0.257696 0.494520
0.326111 0.625491 0.501746
0.995267 0.254346 0.992293
0.160822 0.689054 0.001270
0.995907 0.760753 0.498354
0.159742 0.192300 0.491958
0.811206 0.761223 0.997857
0.325692 0.125479 0.987935
0.656355 0.695175 0.503322
0.656596 0.199917 0.991605
0.487990 0.763078 0.627771
0.845518 0.645378 0.347890
0.623474 0.895186 0.185946
0.616379 0.395875 0.308861
0.093745 0.991831 0.185467
0.092669 0.494591 0.309957
0.847697 0.118765 0.113434
0.475636 0.251449 0.875207
0.327510 0.787484 0.116764
0.720411 0.975740 0.706398
0.200804 0.880147 0.683027
0.975416 0.612789 0.852917
0.986131 0.108285 0.644081
0.204805 0.364607 0.830780
0.718464 0.496262 0.817031
0.323904 0.257705 0.340590
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
BESTgatheredPOSCARS — the same data for the best structure in each generation.
gatheredPOSCARS_unrelaxed — gives all structures produced by USPEX before relaxation.
enthalpies_complete.dat — gives the enthalpies for all structures in each stage of relaxation.
origin — shows which structures originated from which parents and through which variation operators. Example:
ID Origin Enthalpy Parent-E Parent-ID
1 Random -23.395 -23.395 [ 0]
2 Random -23.228 -23.228 [ 0]
3 Random -23.078 -23.078 [ 0]
4 Random -23.195 -23.195 [ 0]
5 Random -23.155 -23.155 [ 0]
6 Random -22.970 -22.970 [ 0]
7 Random -23.131 -23.131 [ 0]
8 Random -23.017 -23.017 [ 0]
9 Random -23.117 -23.117 [ 0]
10 Random -23.195 -23.195 [ 0]
. . . . . . . . . . . . . . . . . . . . . . . . .
gatheredPOSCARS_order — gives the same information as gatheredPOSCARS, and in addition for each atom it gives the values of local order parameters (Ref. 17). Example:
EA1 9.346 8.002 2.688 90.000 90.000 90.000 Sym.group: 1
1.0000
9.346156 0.000000 0.000000
0.000000 8.002181 0.000000
0.000000 0.000000 2.688367
Mg Al O
4 8 16
Direct
0.487956 0.503856 0.516443 1.1399
0.777565 0.007329 0.016443 1.1399
0.987956 0.507329 0.016443 1.1399
0.277565 0.003856 0.516443 1.1399
0.016944 0.178753 0.016443 1.1915
0.019294 0.833730 0.516443 1.2474
0.746227 0.333730 0.516443 1.2474
0.748577 0.678753 0.016443 1.1915
0.516944 0.832431 0.516443 1.1915
0.519294 0.177455 0.016443 1.2474
0.246227 0.677455 0.016443 1.2474
0.248577 0.332431 0.516443 1.1915
0.416676 0.241774 0.516443 1.2914
0.559871 0.674713 0.016443 1.1408
0.205650 0.174713 0.016443 1.1408
0.348845 0.741774 0.516443 1.2914
0.613957 0.380343 0.016443 1.2355
0.804054 0.542164 0.516443 1.2161
0.113957 0.630842 0.516443 1.2355
0.304054 0.469021 0.016443 1.2161
0.848845 0.269411 0.016443 1.2914
0.705650 0.836472 0.516443 1.1408
0.059871 0.336472 0.516443 1.1408
0.916676 0.769411 0.016443 1.2914
0.651564 0.130842 0.516443 1.2355
0.461467 0.969021 0.016443 1.2161
0.151564 0.880343 0.016443 1.2355
0.961467 0.042164 0.516443 1.2161
EA2 9.487 4.757 4.580 90.243 90.188 89.349 Sym.group: 1
1.0000
9.486893 0.000000 0.000000
0.054041 4.756769 0.000000
-0.014991 -0.019246 4.579857
Mg Al O
4 8 16
Direct
0.499837 0.633752 0.011361 0.9368
0.500082 0.131390 0.482012 1.0250
0.813573 0.257696 0.494520 0.9805
0.326111 0.625491 0.501746 0.9437
0.995267 0.254346 0.992293 0.9241
0.160822 0.689054 0.001270 1.1731
0.995907 0.760753 0.498354 0.9696
0.159742 0.192300 0.491958 1.2666
0.811206 0.761223 0.997857 1.0215
0.325692 0.125479 0.987935 1.0353
0.656355 0.695175 0.503322 1.2291
0.656596 0.199917 0.991605 1.2547
0.487990 0.763078 0.627771 1.1199
0.845518 0.645378 0.347890 0.9725
0.623474 0.895186 0.185946 0.9990
0.616379 0.395875 0.308861 1.0141
0.093745 0.991831 0.185467 1.1451
0.092669 0.494591 0.309957 1.1164
0.847697 0.118765 0.113434 0.8821
0.475636 0.251449 0.875207 1.0609
0.327510 0.787484 0.116764 1.0451
0.720411 0.975740 0.706398 0.9539
0.200804 0.880147 0.683027 0.9398
0.975416 0.612789 0.852917 1.1191
0.986131 0.108285 0.644081 0.9803
0.204805 0.364607 0.830780 1.0915
0.718464 0.496262 0.817031 1.0663
0.323904 0.257705 0.340590 1.1471
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
goodStructures_POSCARS and extended_convex_hull_POSCARS (for fixed- and variable-composition calculations correspondingly) report all of the different structures in order of decreasing stability, starting from the most stable structure and ending with the least stable.
compositionStatistics is a file containing statistics of the compositions in terms of which variation operators produced these compositions. Example:
Comp/Ratio Total Random Heredity Mutation Seeds COPEX Best/Convex
[ 0.0000 1.0000 ] 30( 63) 21 8 1 0 0 33
0 8 4( 4) 2 2 0 0 0 0
0 9 2( 2) 2 0 0 0 0 0
0 10 5( 5) 3 2 0 0 0 0
0 11 2( 2) 0 1 1 0 0 0
0 12 6( 35) 6 0 0 0 0 29
0 13 2( 3) 1 1 0 0 0 1
0 14 0( 0) 0 0 0 0 0 0
0 15 2( 2) 2 0 0 0 0 0
0 16 5( 8) 5 0 0 0 0 3
0 3 1( 1) 0 1 0 0 0 0
0 4 1( 1) 0 1 0 0 0 0
[ 0.1250 0.8750 ] 52( 52) 32 9 11 0 0 0
1 7 20( 20) 4 8 8 0 0 0
2 14 32( 32) 28 1 3 0 0 0
[ 0.1111 0.8889 ] 25( 25) 9 14 2 0 0 0
1 8 25( 25) 9 14 2 0 0 0
[ 0.1000 0.9000 ] 16( 16) 9 5 2 0 0 0
1 9 16( 16) 9 5 2 0 0 0
[ 0.0909 0.9091 ] 11( 11) 5 4 2 0 0 0
1 10 11( 11) 5 4 2 0 0 0
[ 0.0833 0.9167 ] 17( 17) 8 2 7 0 0 0
1 11 14( 14) 8 2 4 0 0 0
2 22 3( 3) 0 0 3 0 0 0
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
graphical files (*.pdf) — for rapid visual assessment of the results:
Energy_vs_N.pdf (Fitness_vs_N.pdf) — energy (fitness) as a function of structure number;
Energy_vs_Volume.pdf — energy as a function of volume;
Variation-Operators.pdf — energy of the child vs. parent(s); different operators are marked with different colors (this graph allows one to assess the performance of different variation operators) also show evolution of each operator’s strength.
E_series.pdf — correlation between energies from relaxation steps 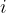 and
and 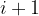 ; helps to detect problems and improve structure relaxation.
; helps to detect problems and improve structure relaxation.
For variable compositions there is an additional graph extendedConvexHull.pdf, which shows the enthalpy of formation as function of composition.
compositionStatistics.pdf — visualization of compositionStatistics file.
Surface_Diagram.pdf — file (only for variable-composition surface structure predictions) showing surface phase diagram.