| USPEX 10.5 manual |
In the field of crystal and cluster structure prediction, several approaches proved to be successful for small systems. Particle Swarm Optimization (PSO), pioneered in this field by Boldyrev 29, is a special class of evolutionary algorithms where a population (swarm) of candidate solutions (called “particles”) is moved in the search space according to a few simple formulae. The movements of the particles are guided by their own best known position in the search space as well as the entire swarm’s best known position. Initially, the coordinates  and ’velocities’
and ’velocities’  of the particles are generated randomly. Then at every step, the positions and velocities are updated according to the formulae:
of the particles are generated randomly. Then at every step, the positions and velocities are updated according to the formulae:
 |
(8) |
Here  ,
,  and
and  are weight factors that control the behavior and efficiency of the PSO algorithm;
are weight factors that control the behavior and efficiency of the PSO algorithm;  and
and  are random numbers in the [0; 1] range generated separately for every particle at every step;
are random numbers in the [0; 1] range generated separately for every particle at every step;  is the best known position of particle
is the best known position of particle 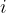 and
and  is the best known position of the entire swarm.
is the best known position of the entire swarm.
Such an algorithm, despite its simplicity, can work 29. Key points to improve with respect to previous implementations 29; 30 are (1) metric of the search space (it is not trivial to map crystal structures uniquely onto a coordinate system) and (2) ways to evolve structures in PSO, i.e. variation operators.
![\includegraphics[scale=0.3]{pic/PSO.png}](images/img-0137.png)
 ,
,  and
and  , respectively.
, respectively.Evolving the particles by determining the speed  (8) directly from coordinates of the atoms and cell parameters of two structures (as in Ref. 30) cannot be productive. Our solution is to use fingerprint distances 17 as the most natural metric for the energy landscape, and variation operators of USPEX for evolving the ’PSO particles’ (i.e. structures) as the most efficient unbiased ways to evolve a population of structures. Namely, the particle is either mutated (to imitate a random move), or participates in heredity with its best known position or in heredity with the best known population position (to imitate PSO moves in the direction of these positions). Instead of applying at each step all moves with some weights (see Eq. 8), we apply them one at a time with probabilities described by formulae:
(8) directly from coordinates of the atoms and cell parameters of two structures (as in Ref. 30) cannot be productive. Our solution is to use fingerprint distances 17 as the most natural metric for the energy landscape, and variation operators of USPEX for evolving the ’PSO particles’ (i.e. structures) as the most efficient unbiased ways to evolve a population of structures. Namely, the particle is either mutated (to imitate a random move), or participates in heredity with its best known position or in heredity with the best known population position (to imitate PSO moves in the direction of these positions). Instead of applying at each step all moves with some weights (see Eq. 8), we apply them one at a time with probabilities described by formulae:
 |
(11) |
where  is a fingerprint distance between current and best position of a particle, while
is a fingerprint distance between current and best position of a particle, while  is a fingerprint distance between the current position of the particle and best known position of the entire population. Our tests, performed on a few diverse systems, show that this approach (which we call “cor-PSO”, i.e. corrected PSO) is relatively successful and works better than previous versions of PSO, but still cannot compete with the USPEX algorithm 2; 31 for success rate or efficiency.
is a fingerprint distance between the current position of the particle and best known position of the entire population. Our tests, performed on a few diverse systems, show that this approach (which we call “cor-PSO”, i.e. corrected PSO) is relatively successful and works better than previous versions of PSO, but still cannot compete with the USPEX algorithm 2; 31 for success rate or efficiency.
The following variables are unique for calculationMethod=PSO:
 variable blue PSO_softMut
variable blue PSO_softMut
Meaning: Weight of softmutation ( in eq. 11).
in eq. 11).
Default: 1
Format:
1 : PSO_softMut
 variable blue PSO_BestStruc
variable blue PSO_BestStruc
Meaning: Weight of heredity with the best position of the same PSO particle ( in eq. 11).
in eq. 11).
Default: 1
Format:
1 : PSO_BestStruc
 variable blue PSO_BestEver
variable blue PSO_BestEver
Meaning: Weight of heredity with the globally best PSO particle ( in eq. 11).
in eq. 11).
Default: 1
Format:
1 : PSO_BestEver