| USPEX 10.5 manual |
To predict surface reconstructions, you have to:
Specify 200 or 201 : calculationType.
Provide a file containing substrate in VASP5 POSCAR format, as shown on Fig. 12. This can be done using online utility: https://uspex-team.org/online_utilities/substrate/ (after you get your slab model, you need to put it in your folder and run USPEX one time — it will generate for you the "final" POSCAR_SUBSTRATE file at stop. With this file you can now run your USPEX calculations).
Do not forget to specify vacuumSize which should be sufficently large to prevent interaction between periodic images of slabs.
Specify the following parameters:
![\includegraphics[scale=0.5]{pic/Surface.png}](images/img-0110.png)
% symmetries 2-17 % endSymmetries
Note: If the symmetries tag is present, USPEX will try to generate surface structures using plane groups.
 variable thicknessS
variable thicknessS
Meaning: Thickness of surface region. Adatoms are allowed only in this region.
Default: 2.0 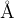
Format:
3.5 : thicknessS
 variable thicknessB
variable thicknessB
Meaning: Thickness of buffer region in substrate. This region is part of POSCAR_SUBSTRATE, and is allowed to relax.
Default: 3.0 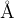
Format:
3.0 : thicknessB
 variable reconstruct
variable reconstruct
Meaning: Maximum multiplications of the surface cell, to allow for complex reconstructions.
Default: 1
Format:
1 : reconstruct
USPEX is capable of predicting surface reconstructions with variable number of surface atoms. In this case, stable surface reconstructions are dictated by chemical potentials 28. A typical set of input is the following:
****************************************** * TYPE OF RUN AND SYSTEM * ****************************************** USPEX : calculationMethod (USPEX, VCNEB, META) 201 : calculationType (dimension: 0-3; molecule: 0/1; varcomp: 0/1) % atomType Si O % EndAtomType % numSpecies 2 4 % EndNumSpecies
Here we specify the maximum number of surface atoms in a  cell.
cell.
****************************************** * SURFACES * ****************************************** % symmetries 2-17 % endSymmetries % StoichiometryStart 1 2 % StoichiometryEnd
This defines the stoichiometry of the bulk.
-23.7563 : E_AB (DFT energy of AmBn compound, in eV per formula unit) -5.4254 : Mu_A (DFT energy of elemental A, in eV/atom) -4.9300 : Mu_B (DFT energy of elemental B, in eV/atom) 3.5 : thicknessS (thickness of surface region, 2 A by default ) 3.0 : thicknessB (thickness of buffer region in substrate, 3 A by default) 4 : reconstruct (maximum multiplications of cell)
At the moment, USPEX supports variable-composition calculation for the following cases:
Reconstructions of elemental surfaces (such as C@diamond(100) surface).
Reconstructions of surfaces at binary compounds (such as GaN(0001) surface).
Reconstructions involving foreign species on elemental surfaces (such as PdO@Pd(100) surface).