| USPEX 10.2 manual |
Transition path sampling (TPS) is a method for simulating rare events: a good example of a rare event is a transition of a system from one stable state to another; these occur due to rare fluctuations, which cannot be observed within typical timescales of molecular dynamics simulations. Examples include protein folding, chemical reactions and nucleation. TPS is a powerful method to study these phenomena. In short, TPS is a technique performing Monte-Carlo sampling over transition trajectories (each of which is a molecular dynamics trajectory).
The TPS method is only enabled with the LAMMPS and CP2K (will be available soon) codes at the moment.
To switch on the TPS mode, you have to:
Specify
TPS : calculationMethod
Create a MD restart file lammps.restart for LAMMPS code or cp2k.restart for CP2K code in the Seeds/ folder.
Create an order parameter calculation script or use USPEX’s fingerprints as default. (See more details below).
Specify the following TPS options:
 variable numIterations
variable numIterations
Meaning: Maximum number of TPS steps.
Default: 1000
Format:
500 : numIterations
 variable speciesSymbol
variable speciesSymbol
Meaning: Describes identities of all chemical species (atomic or molecular).
Default: No default
Format:
% speciesSymbol
CH4 Si O
% EndSpeciesSymbol
Note: In TPS, we use speciesSymbol to replace the atomType to support molecular species, together with numSpecies. For example, we can use
% numSpecies
1 648 1296
% EndNumSpecies
% speciesSymbol
CH4 Si O
% EndSpeciesSymbol
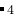 ) molecule in a silica zeolite (SiO
) molecule in a silica zeolite (SiO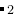 ). In this case, methane molecule will be considered as a whole particle in writing MD input files in LAMMPS.
). In this case, methane molecule will be considered as a whole particle in writing MD input files in LAMMPS.  variable mass
variable mass
Meaning: Masses for each type of species.
Default: Default value corresponding to the species
Format:
% mass
16.000
% EndMass
 variable amplitudeShoot
variable amplitudeShoot
Meaning: Distribution amplitude of the momentum for A B and B
B and B A directions in shooting operation.
A directions in shooting operation.
Default: 0.1 0.1
Format:
% amplitudeShoot
0.12 0.12
% EndAmplitudeShoot
 variable magnitudeShoot
variable magnitudeShoot
Meaning: Factor for increasing or decreasing the amplitude of magnitude distribution when succeeded or failed to have a new MD trajectory, respectively.
Default: 1.05 1.05
Format:
% magnitudeShoot
1.0 1.05
% EndMagnitudeShoot
Note: Unless the user has system-specific experience, we strongly suggest to use this option to automatically adjust the distribution amplitude of the momentum.
 variable shiftRatio
variable shiftRatio
Meaning: Ratio of applying a shifter operation after having a successful shooter operation.
Default: 0.1
Format:
0.5 : shiftRatio
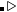 variable orderParaType
variable orderParaType
Meaning: Method of order parameter calculation to distinguish different phases:
“0” — user-defined method to calculate the order parameter, a script should be provided in cmdOrderParameter;
“1” — fingerprint method (Oganov & Valle, 2009).
Default: No default
Format:
0 : orderParaType
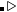 variable opCriteria
variable opCriteria
Meaning: Two values here specify the tolerable degree of similarity to the starting and ending states, respectively. If one or both ends of the trajectory lead to different end structures, such a trajectory will be rejected in the TPS method.
Default: No default
Format:
% opCriteria
0.995 0.995
% EndOpCriteria
Note: If you use the fingerprint method, the larger system you have, the more strict criteria you should have. For example, we should set opCriteria=[ 0.995, 0.995 ] with 1,000 atoms; and opCriteria=[ 0.998, 0.998 ] in system more than 30,000 atoms when studying the fcc hcp phase transition.
hcp phase transition.
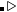 variable cmdOrderParameter
variable cmdOrderParameter
Meaning: User-defined command to calculate the order parameter. It is not needed if you use the fingerprint method.
Default: No default
Format:
% cmdOrderParameter
./extractOp.sh
% EndCmdOrderParameter
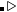 variable cmdEnthalpyTemperature
variable cmdEnthalpyTemperature
Meaning: User-defined command to extract the enthalpy and temperature from MD results.
Default: No default
Format:
% cmdEnthalpyTemperature
./extractHT.sh
% EndCmdEnthalpyTemperature
 variable orderParameterFile
variable orderParameterFile
Meaning: File name to store the order parameter history in a single MD calculation for TPS to read.
Default: fp.dat
Format:
op.dat : orderParameterFile
 variable enthalpyTemperatureFile
variable enthalpyTemperatureFile
Meaning: File name to store the enthalpy & temperature history in a single MD calculation for TPS to read or user-defined script to analyze.
Default: HT.dat
Format:
HT0.dat : enthalpyTemperatureFile
 variable trajectoryFile
variable trajectoryFile
Meaning: File name to store the MD trajectory. This name should be consistent with the MD trajectory output file name from the calculation.
Default: traj.dat
Format:
trajectory.xyz : trajectoryFile
 variable MDrestartFile
variable MDrestartFile
Meaning: File name to store the MD restart file from the calculation for TPS to read. This name should be consistent with the MD restart file name from the calculation.
Default: traj.restart
Format:
lammps0.restart : MDrestartFile