| USPEX 10.5 manual |
 variable blue mutationRate
variable blue mutationRate
Meaning: Standard deviation of the strain matrix components for lattice mutation. The strain matrix components are selected randomly from the Gaussian distribution and are only allowed to take values between -1 and 1. Lattice mutation essentially incorporates the ideas of metadynamics into our method 6; 25, where new structures are found by building up cell distortions of some known structure. Unlike in metadynamics, the distortions are not accumulated in our method, so the strain components should be large enough to obtain new structures.
Default: 0.5
Format:
0.5 : mutationRate
It is a good idea to combine lattice mutation with a weak softmutation:
 variable blue mutationDegree
variable blue mutationDegree
Meaning: The maximum displacement in softmutation in 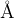 . The displacement vectors for softmutation or coordinate mutation are scaled so that the largest displacement magnitude equals mutationDegree.
. The displacement vectors for softmutation or coordinate mutation are scaled so that the largest displacement magnitude equals mutationDegree.
Default: 3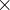 (average atomic radius)
(average atomic radius)
Format:
2.5 : mutationDegree
 variable blue ordering_active
variable blue ordering_active
Meaning: Switch on the biasing of variation operators by local order parameters.
Default: 1
Format:
1 : ordering_active
 variable blue symmetrize
variable blue symmetrize
Meaning: Switches on a transformation of all structures to standard symmetry-adapted crystallographic settings.
Default: 0
Format:
1 : symmetrize
 variable blue valenceElectr
variable blue valenceElectr
Meaning: Number of valence electrons for each type of atoms.
Default: these numbers are constants for all atoms, and we have tabulated them, no need to specify explicitly.
Format:
% valenceElectr
2 6
% EndValenceElectr
 variable blue percSliceShift
variable blue percSliceShift
Meaning: Probability of shifting slabs (used in heredity) in all dimensions, 1.0 means 100%.
Default: 1.0
Format:
0.5 : percSliceShift
 variable blue maxDistHeredity
variable blue maxDistHeredity
Meaning: Specifies the maximal fingerprint cosine distances between structures that participate in heredity. This specifies the radius on the landscape within which structures can mate. Use with care (or do not use at all).
Default: 0.5
Format:
0.5 : maxDistHeredity
 variable blue manyParents
variable blue manyParents
Meaning: Specifies whether more than two slices (or more than two parent structures) should be used for heredity. This may be beneficial for very large systems.
Possible values (integer):
0 — only 2 parents are used, 1 slice each.
1 — many structures are used as parents, 1 slice each.
2 — two structures are used as parents, many slices (determined dynamically using parameters minSlice and maxSlice) are chosen independently from each one.
3 — two structures are used as parents, many slices (determined dynamically using parameters minSlice and maxSlice) are cut from the cell with a fixed offset. This is the preferred option for large systems. For example, we cut both structures into slices of approximately the same thickness and then choose the even slices from parent 1 and odd slices form parent 2, making a multilayered “sandwich”, or a “zebra”.
Default: 0
Format:
3 : manyParents
blue minSlice, maxSlice: Determines the minimal and maximal thickness of the slices in 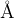 that will be cut out of the parent structures to participate in creation of the child structure. We want the slices to be thick enough to carry some information about the parent (but not too thick to make heredity ineffective). Reasonable values for these parameters are around 1 and 6
that will be cut out of the parent structures to participate in creation of the child structure. We want the slices to be thick enough to carry some information about the parent (but not too thick to make heredity ineffective). Reasonable values for these parameters are around 1 and 6 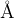 , respectively.
, respectively.
For clusters, you can directly specify the number of parents participating in heredity (but we found this to be of little use):
 variable blue numberparents
variable blue numberparents
Meaning: Defines the number of parents in heredity for clusters.
Default: 2
Format:
2 : numberparents