| USPEX 10.5 manual |
Once you have downloaded the USPEX package and installed it, you can run the first USPEX example. The description of the examples is listed in Appendix 9.1. The required external codes to run the examples (except EX13) are shown below:
GULP: EX02, EX03, EX08, EX12, EX15 (VCNEB), EX16, EX18, EX21(META), EX22(Generalized META), EX23 VASP: EX01, EX07, EX09, EX14 (META), EX17, EX19, EX24, EX29, EX30, EX31 QE: EX33 LAMMPS: EX04, EX26 (TPS) ATK: EX05 CASTEP: EX06 DMACRYS: EX10 Tinker: EX11 MOPAC: EX20 DFTB: EX25 FHIaims: EX27, EX28
Now, let us start our first USPEX experience:
To get the version information, you can use the following command:
>> USPEX -v
You should get something like this:
USPEX Version 10.2 (19/01/2019)
EX13 does not require any external code, you can run it to get familiar with the USPEX running procedure. The calculation would take  30 minutes. To start the calculation, first create a test folder, copy the example files and then run the calculation through the USPEX Python runner, with the following commands:
30 minutes. To start the calculation, first create a test folder, copy the example files and then run the calculation through the USPEX Python runner, with the following commands:
>> mkdir EX13
>> cd EX13
>> USPEX -c 13
>> USPEX -r
Meanwhile you have some time to get more details about example EX13. In EX13, the structural order is optimized (minimized) with the evolutionary algorithm to yield a generalized special quasirandom structure (gSQS). Therefore in INPUT.txt the following is set:
USPEX : calculationMethod
% optType
-4
% EndOptType
and these parameters are used below:
300 : calculationType
% atomType
Co Ti O
% EndAtomType
% numSpecies
16 16 64
% EndNumSpecies
which specifies that we deal with the Co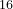 Ti
Ti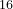 O
O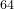 system.
system.
Since you do not need to use any external code, you can simply set
% abinitioCode
0
% ENDabinit
The Seeds/POSCARS file contains the initial Ti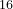 Co
Co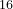 O
O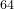 structure.
structure.
When you find the USPEX_IS_DONE file, congratulations, you have successfully finished your first USPEX example. Next, you can run calculations which require external codes.
In this step, we suggest to run examples interfaced with GULP or VASP, starting from EX02 or EX01. Use USPEX runner to get the example information of EX02, create a separate folder and copy the files, with commands:
>> mkdir EX02
>> cd EX02
>> USPEX -c 2
Since in example EX02 GULP code is used, set:
% abinitioCode
3 3 3 3
% ENDabinit
To run a serial job without a job batch system, you should change the following parameters in the INPUT.txt below:
1 : whichCluster
1 : numParallelCalcs
In the example INPUT.txt, you will see whichCluster=QSH, where QSH is name of our group’s cluster name. User can specify their own cluster: for the details, please see Section 8.8.
% commandExecutable
gulp < input > output
% EndExecutable
But make sure, that this command also works on your machine. If you want to run EX01 with a parallel version of VASP, you should set something like:
% abinitioCode
1 1 1 1
% ENDabinit
% commandExecutable
mpirun -np 8 vasp
% EndExecutable
With wrong commandExecutable settings, you will not able to run your calculation. Finally, do not forget to suitably modify submitJob_remote.py and checkStatus_remote.py files for your computer/supercomputer.
When everything is set correctly, we can run the calculation through USPEX runner using the command:
>> USPEX -r
After starting the command, you can check the output files in results1/ folder.
Now, you have a basic experience of using USPEX to run simple calculations. Please read the following sections of this manual to get more insight into USPEX. When analyzing results, it is essential that you visualize the structures (for visualization, see section 8.1).