| USPEX 10.5 manual |
USPEX employs a powerful two-level parallelization scheme, making its parallel scalability exemplary. The first level of parallelization is performed within structure relaxation codes, the second level of parallelization distributes the calculation over the individuals in the same population (since structures within the same generation are independent of each other).
First, you must specify which code(s) you want to use for structure relaxation and fitness calculation:
 variable blue abinitioCode
variable blue abinitioCode
Meaning: Defines the code used for every optimization step.
Default: 1 for every optimization step (VASP)
Format:
% abinitioCode
3 2 2 1 1
% ENDabinit
Alternative Format:
% abinitioCode
1 1 1 1 1 (14)
% ENDabinit
The difference is that here one (or more) stage(s) of calculation for each structure is shown in parentheses. This is very useful when optimizing physical properties: for calculations in parentheses, the structure is symmetrized and represented in the standard crystallographic setting; this allows us to fully use crystal symmetry and make property calculations cheaper and more numerically robust. Before symmetrization and property calculations, the structure must be well relaxed (so the user must make sure that relaxation is well done).
Note 1: the enthalpy is taken from the last stage BEFORE parentheses.
Note 2: Numbers indicate the code used at each step of structure relaxation:
1 — VASP
2 — SIESTA
3 — GULP
4 — LAMMPS
5 — ORCA
6 — DMACRYS
7 — CP2K
8 — Quantum Espresso
9 — FHI-aims
10 — ATK
11 — CASTEP
12 — Tinker
13 — MOPAC
14 — BoltzTraP
15 — DFTB
16 — Gaussian
(only for clusters)
18 — Abinit
19 — CRYSTAL
 variable blue KresolStart
variable blue KresolStart
Meaning: Specifies the reciprocal-space resolution for 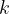 -points generation (units:
-points generation (units: 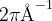 ).
).
Default: from 0.2 to 0.08 linearly
Format:
% KresolStart
0.2 0.16 0.12 0.08
% Kresolend
NOTE: You can enter several values (one for each step of structure relaxation), starting with cruder (i.e., larger) values and ending with high resolution. This dramatically speeds up calculations, especially for metals, where very many k-points are needed. This keyblock is important for ab-initio calculations (through some codes, e.g. VASP and SIESTA, now have similar tricks)).
For clusters, 2D-crystals, and surfaces, you have to specify the thickness of the vacuum region around the cluster (or around the surface slab):
 variable blue vacuumSize
variable blue vacuumSize
Meaning: Defines the amount of vacuum added around the structure (closest distance in 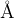 between neighboring clusters in adjacent unit cells). Used only for surfaces, 2D-crystals, and nanoparticles.
between neighboring clusters in adjacent unit cells). Used only for surfaces, 2D-crystals, and nanoparticles.
Default: 10 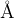 for every step of relaxation
for every step of relaxation
Format:
% vacuumSize
10 10 15 20 20
% EndVacuumSize
 variable blue numParallelCalcs
variable blue numParallelCalcs
Meaning: Specifies how many structure relaxations you want to run in parallel.
Default: 1
Format:
10 : numParallelCalcs
You need to supply the job submission files or the names of executable files for each code/mode you are using.
 variable blue commandExecutable
variable blue commandExecutable
Meaning: Specifies the name of the job submission files or executables for a given code.
Default: no default, has to be specified by the user.
Format:
% commandExecutable
gulp < input > output
mpirun -np 8 vasp > out
mpirun -np 8 vasp > out
mpirun -np 8 vasp > out
% EndExecutable
NOTE: Every line corresponds to a stage of relaxation — the first line describes the execution of the first stage of relaxation, etc. For example, abinitioCode equal to “3 1 1 1” means that the first relaxation step will be performed with GULP, while the subsequent steps will be performed using VASP via the command “mpirun -np 8 vasp > out”. If only one line is present in commandExecutable, then the same execution will be performed for all steps of relaxation.
You can actually use USPEX on practically any platform in the remote submission mode. In that case, your workstation will prepare input (including jobs), send them to the remote compute nodes, check when the calculations are complete, get the results back, analyze them, and prepare new input. The amount of data being sent to and fro is not large, so the network does not need to be very fast. Job submission is, of course, machine-dependent.
 variable blue whichCluster
variable blue whichCluster
Meaning: Specifies the types of job submission.
Possible values (integer):
0 — no-job-script;
1 — local submission;
2 — remote submission.
Default: 0
Format:
1 : whichCluster
 variable blue remoteFolder
variable blue remoteFolder
Meaning: Folder on the remote supercomputer where the calculation will be performed. This keyword is activated only if whichCluster=2.
Default: none
Format:
Blind_test : remoteFolder
NOTE: there is a similar parameter specified in the remote submission file — homeFolder. The actual path to the calculation will be /*remoteFolder*/CalcFolderX where X=1, 2, 3,….
 variable blue PhaseDiagram
variable blue PhaseDiagram
Meaning: Enables calculation of a phase diagram for calculationType=300 and 301. It gives an idea (crude one — just to get a rough idea!) of which structures may be stable at higher and lower pressures, and a rough idea of transition pressures.
Default: 0
Format:
1 : PhaseDiagram