| USPEX 9.4.4 manual |
A family of antiseed techniques have been developed and implemented in USPEX, all based on the idea of penalizing already sampled structures to ensure that the simulation is not stuck in a local minimum. Here, time-dependent fitness is the sum of the actual enthalpy (or another fitness property of interest) and a history-dependent term, which is the sum of the Gaussian potentials added to already sampled parts of the energy landscape:
![\[ f=f_0 + \sum _{a} W_ a \exp \left(-\frac{d_{ia}^{2}}{2\sigma _{a}^{2}}\right), \]](images/img-0083.png) |
where 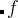 is fitness (
is fitness (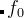 — the true fitness,
— the true fitness, 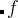 — history-dependent fitness),
— history-dependent fitness), 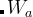 is the height and
is the height and 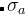 is the width of the Gaussian. In our approach, Gaussian parameters change depending on the population diversity and energy spread at each generation.
is the width of the Gaussian. In our approach, Gaussian parameters change depending on the population diversity and energy spread at each generation.
There are three ways to use this technique. In the first, you can put the structure that you wish to penalize in the AntiSeeds folder. For example, this can be the ground state structure — in this case, USPEX will try to find the second lowest-enthalpy structure.
In the second and third methods, you don’t specify antiseed structure(s) — the calculation either uses all sampled structures as antiseeds (well tested; the recommended approach) or just the best structure in each generation. You need to specify a few settings:
 variable antiSeedsActivation
variable antiSeedsActivation
Meaning: Specifies from which generation the antiseed mode will be switched on. When antiSeedsActivation 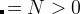 , Gaussians are added to all structures starting from generation
, Gaussians are added to all structures starting from generation 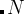 , and when
, and when 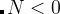 — Gaussians are only added to the best structure of each generation, starting from generation
— Gaussians are only added to the best structure of each generation, starting from generation 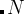 . When
. When 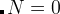 , Gaussians are only added to the structures put in the AntiSeeds folder. If you don’t want to use antiseeds, specify very large antiSeedsActivation (for example, 5000) and antiSeedsMax=0.0.
, Gaussians are only added to the structures put in the AntiSeeds folder. If you don’t want to use antiseeds, specify very large antiSeedsActivation (for example, 5000) and antiSeedsMax=0.0.
Default: 5000
Format:
1 : antiSeedsActivation
 variable antiSeedsMax
variable antiSeedsMax
Meaning: Specifies the height of the Gaussian, in units of the mean square deviation of the enthalpy in the generation (computed only among bestFrac structures, i.e., among potential parents). We recommend antiSeedsMax=0.01.
Default: 0.000
Format:
0.005 : antiSeedsMax
 variable antiSeedsSigma
variable antiSeedsSigma
Meaning: Specifies the width of the Gaussian, in units of the average distance between structures in the generation (computed only among bestFrac structures, i.e., among potential parents). We recommend antiSeedsSigma=0.005.
Default: 0.001
Format:
0.005 : antiSeedsSigma
Fig. 7 shows an example of use of antiseed technique.
![\includegraphics[scale=0.3]{pic/antiseeds}](images/img-0091.png)